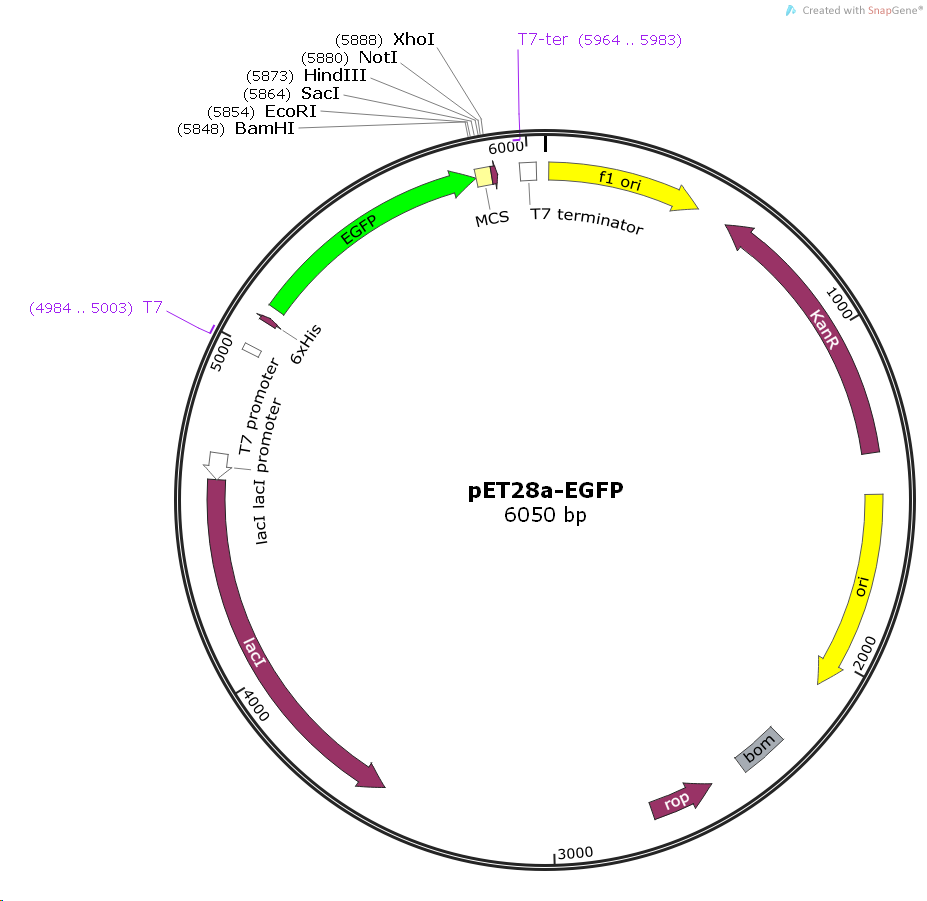
pET28a-EGFP
Search name
pET28a-EGFP,Plasmid pET28a-EGFP,pET28a-EGFP vector
Bacterial Resistance:Kanamycin
Growth Strain:DH5α
Expression: Bacterial
Use:pET Plasmid
Promoter: T7/lac
Replicator: ColE1, ori, F1, ori
Terminator: T7 Terminator
Plasmid classification: large intestine series plasmid, large intestine fluorescent plasmid, large intestine green plasmid
Plasmid size: 6050bp
Plasmid labels: N-6 * His, N-Thrombin, N-EGFP, C-6 * His
Prokaryotic resistance: kanamycin Kan (50 g/ml)
Clone strain: DH5 alpha and other Escherichia coli
Culture conditions: 37 DEG C, aerobic, LB
Expressing host: BL21 (DE3) and other E. coli
Culture conditions: 37 DEG C, aerobic, LB
Induction: IPTG or lactose and its analogues
Primers for 5'sequencing: T7 (TAATACGACTCACTATAGGG)
Primers for 3'sequencing: T7-ter (TGCTAGTTATTGCTCAGCGG)
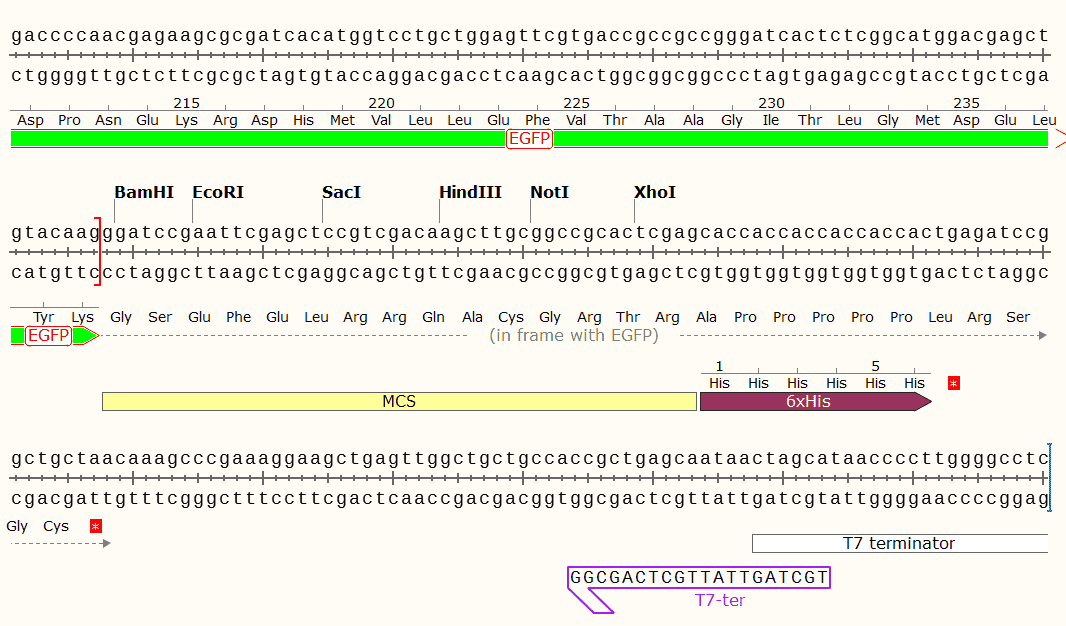
Sequence
LOCUS Exported 6050 bp ds-DNA circular SYN 12-JUL-2017
DEFINITION synthetic circular DNA
KEYWORDS pET28a-EGFP
SOURCE synthetic DNA construct
ORGANISM synthetic DNA construct
REFERENCE 1 (bases 1 to 6050)
TITLE Direct Submission
JOURNAL Exported 2015-4-17
REFERENCE 2 (bases 1 to 6050)
AUTHORS .
TITLE Direct Submission
JOURNAL Exported Wednesday, July 12, 2017
FEATURES Location/Qualifiers
source 1..6050
/organism="synthetic DNA construct"
/mol_type="other DNA"
rep_origin 12..467
/direction=RIGHT
/note="f1 ori"
/note="f1 ori
f1 bacteriophage origin of replication; arrow indicates
direction of (+) strand synthesis"
CDS complement(560..1375)
/codon_start=1
/gene="aph(3')-Ia"
/product="aminoglycoside phosphotransferase"
/note="aph(3')-Ia"
/note="KanR
confers resistance to kanamycin in bacteria or G418
(Geneticin(R)) in eukaryotes"
/translation="MSHIQRETSCSRPRLNSNMDADLYGYKWARDNVGQSGATIYRLYG
KPDAPELFLKHGKGSVANDVTDEMVRLNWLTEFMPLPTIKHFIRTPDDAWLLTTAIPGK
TAFQVLEEYPDSGENIVDALAVFLRRLHSIPVCNCPFNSDRVFRLAQAQSRMNNGLVDA
SDFDDERNGWPVEQVWKEMHKLLPFSPDSVVTHGDFSLDNLIFDEGKLIGCIDVGRVGI
ADRYQDLAILWNCLGEFSPSLQKRLFQKYGIDNPDMNKLQFHLMLDEFF"
rep_origin 1497..2085
/direction=RIGHT
/note="ori"
/note="ori
high-copy-number ColE1/pMB1/pBR322/pUC origin of
replication"
misc_feature 2271..2413
/note="bom"
/note="bom
basis of mobility region from pBR322"
CDS complement(2515..2706)
/codon_start=1
/gene="rop"
/product="Rop protein, which maintains plasmids at low copy
number"
/note="rop"
/translation="MTKQEKTALNMARFIRSQTLTLLEKLNELDADEQADICESLHDHA
DELYRSCLARFGDDGENL"
CDS complement(3515..4597)
/codon_start=1
/gene="lacI"
/product="lac repressor"
/note="lacI"
/note="lacI
The lac repressor binds to the lac operator to inhibit
transcription in E. coli. This inhibition can be relieved
by adding lactose or isopropyl-beta-D-thiogalactopyranoside
(IPTG)."
/translation="MKPVTLYDVAEYAGVSYQTVSRVVNQASHVSAKTREKVEAAMAEL
NYIPNRVAQQLAGKQSLLIGVATSSLALHAPSQIVAAIKSRADQLGASVVVSMVERSGV
EACKAAVHNLLAQRVSGLIINYPLDDQDAIAVEAACTNVPALFLDVSDQTPINSIIFSH
EDGTRLGVEHLVALGHQQIALLAGPLSSVSARLRLAGWHKYLTRNQIQPIAEREGDWSA
MSGFQQTMQMLNEGIVPTAMLVANDQMALGAMRAITESGLRVGADISVVGYDDTEDSSC
YIPPLTTIKQDFRLLGQTSVDRLLQLSQGQAVKGNQLLPVSLVKRKTTLAPNTQTASPR
ALADSLMQLARQVSRLESGQ"
promoter complement(4598..4675)
/gene="lacI"
/note="lacI lacI promoter"
/note="lacI promoter"
promoter 4984..5002
/note="T7 promoter"
/note="T7 promoter
promoter for bacteriophage T7 RNA polymerase"
CDS 5083..5100
/codon_start=1
/product="6xHis affinity tag"
/note="6xHis affinity tag"
/note="6xHis"
/translation="HHHHHH"
CDS 5131..5847
/codon_start=1
/product="enhanced GFP"
/note="enhanced GFP"
/note="EGFP
mammalian codon-optimized"
/translation="MVSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTL
KFICTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTIFFKDD
GNYKTRAEVKFEGDTLVNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIK
VNFKIRHNIEDGSVQLADHYQQNTPIGDGPVLLPDNHYLSTQSALSKDPNEKRDHMVLL
EFVTAAGITLGMDELYK"
misc_feature 5848..5893
/note="MCS"
CDS 5894..5911
/codon_start=1
/product="6xHis affinity tag"
/note="6xHis affinity tag"
/note="6xHis"